Set II. RNA-seq analysis of NHP tissue-specific transcriptomes
Tissue-specific RNA-seq was performed for 11 of the original 15 NHP species/subspecies from ~15 different tissues. We have made this data, the first large collection of tissue-specific primate transcriptomic data, pubically available. This data and further information concerning it is published in Nucleic Acid Research and can be found here.
Raw Sequence Data
The complete tissue-specific RNA-seq dataset consists of 157 libraries across 14 species/subspecies. The raw data sets contain over 10 billion read pairs (100x100bp) totaling to 2.44 terabases of Illumina sequence. A summary of the amount of tissue-specific raw RNA-seq data can be found on Figure 1.
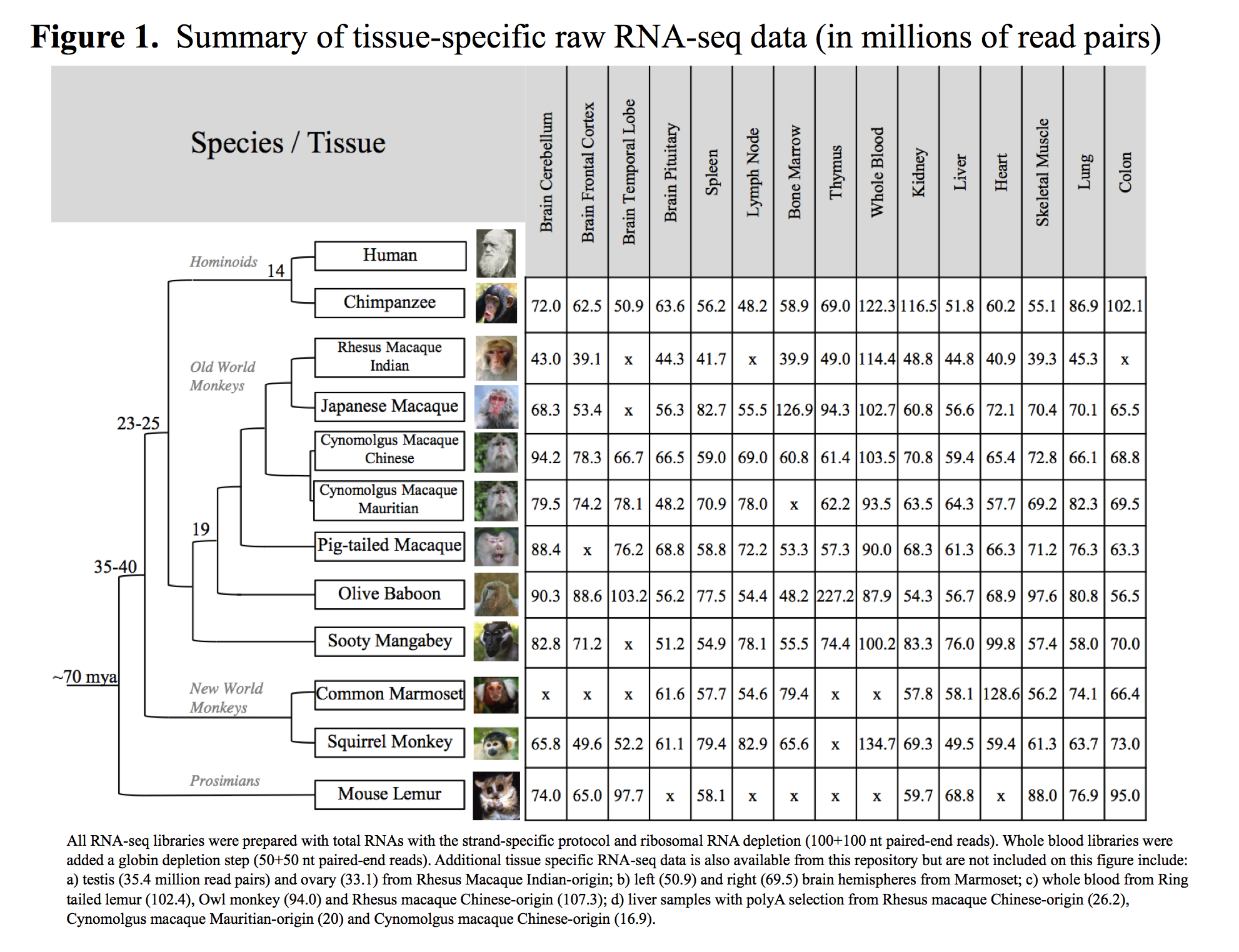
On average, there are 66.8 million paired-end reads per tissue and species for non-whole blood samples (ranging from 42 million on average in RMI to 95 millions in chimpanzee) and around 100 million paired-end reads for whole blood samples.
Tissue-specific RNA-seq analysis for Indian-origin rhesus macaque was performed by the Division of NHP Systems Biology at the Washington National Primate Research Center at the University of Washington.
Dr. Gary Schroth’s laboratory at Illumina generated tissue-specific total RNA-seq sequencing libraries for the other NHP species and tissues. These libraries were provided to the NHP Genomics Group (headed by Dr. Jeffrey Rogers) at the Baylor Human Genome Sequencing Center for HiSeq sequencing.
To address the particular technical challenge of extremely abundant globin transcripts in whole blood, Dr. Schroth applied a special protocol to prepare total RNA-seq sequencing libraries that were depleted of ribosomal RNA and globin transcripts. His laboratory then performed the sequencing, generating an average of ~100 million 2 x 50 nt paired-end reads per library.
To access Set II data click here to enter your contact information. Your information is not shared and is used only to track the number of investigators using our resources for reporting to funding agencies.
Tissue-specific Expression Abundance
The sequence quality of the raw reads is such that 88% of the reads were aligned to human reference sequences, allowing us to compute the full list of expression abundance across all tissues for every species (using both Refseq and Aceview).
The correct assignment of tissues was verified using the covariance analysis of the expression patterns provided by the Magic pipeline. Examples of some tissue-specific genes can be seen in Figure 2.
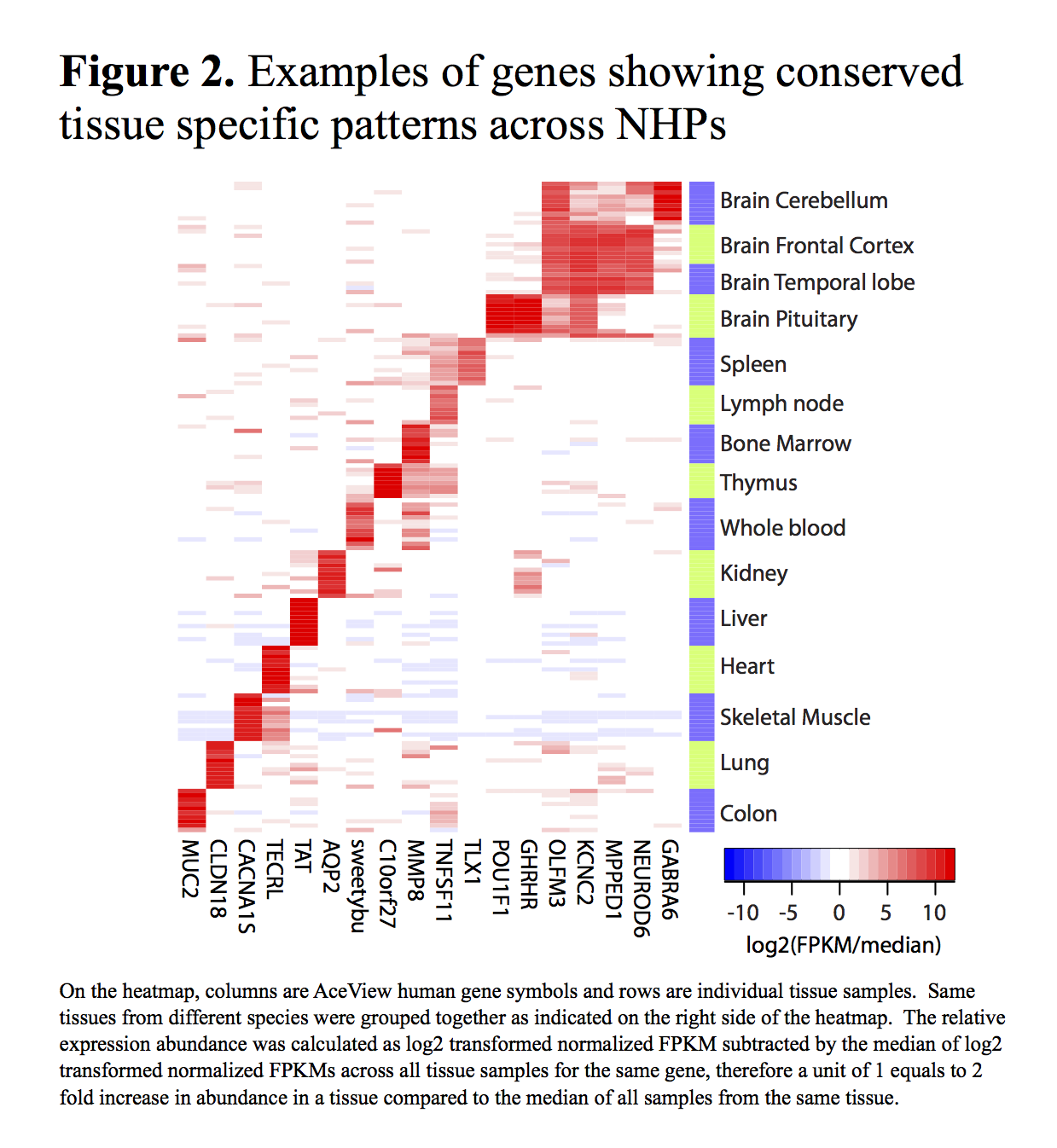
The full expression data matrix has also been made available through this website.
To access Set II data click here to enter your contact information. Your information is not shared and is used only to track the number of investigators using our resources for reporting to funding agencies.
AceView
AceView provides a curated, comprehensive and non-redundant sequence representation of all public mRNA sequences (mRNAs from GenBank or RefSeq, and single pass cDNA sequences from dbEST and Trace). These experimental cDNA sequences are first co-aligned on the genome then clustered into a minimal number of alternative transcript variants and grouped into genes. Using exhaustively and with high quality standards the available cDNA sequences evidences the beauty and complexity of mammals’ transcriptome, and the relative simplicity of the nematode and plant transcriptomes. Genes are classified according to their inferred coding potential; many presumably non-coding genes are discovered. Genes are named by Entrez Gene names when available, else by AceView gene names, stable from release to release.
In addition to the complete full expression data matrix downloadable on this website, all of this information is also integrated into the AceView databases. Each species' tissue-specific reads have been aligned to a human reference. The AceView website provides users with an interfaces to browse through this human reference for individual genes of interest. For each gene, expression levels and conservation across all tissues and species from the NHPRTR are provided.
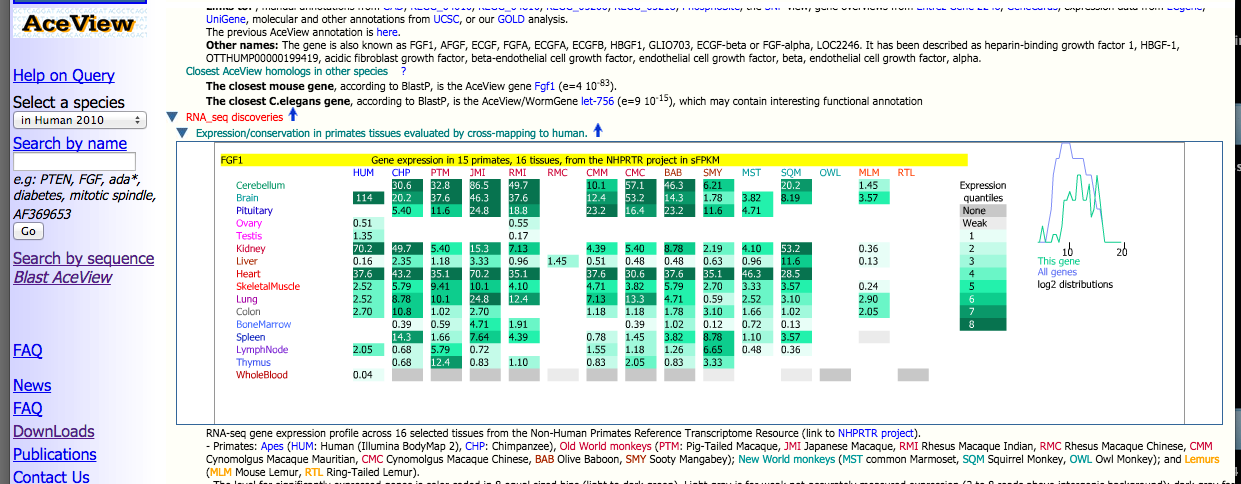
Above is a glimpse of the results a user can retrieve from AceView through searching the human reference by gene name (FGF1 in this example). An expression profile for the gene in all primates and tissues is provided amongst a wide variety of other information.
