Set IV. Full-length Indian-origin rhesus macaque transcriptome using PacBio Iso-Seq technology
Using PacBio Iso-Seq, we generated over 2.8 million transcript sequencing reads (Circular Consensus Sequence reads, CCSs), ranging from 300 to 45,549 nucleotides, from Indian-origin rhesus macaque lymph node tissue, rectal tissue, PBMC and whole blood. For comparative analysis of splicing patterns against existing annotation, these CCSs were processed and aligned to both rhesus macaque and human reference genomes. Using Iso-Seq and its supporting Cupcake scripts (https://github.com/Magdoll/cDNA_Cupcake), aligned CCSs were processed into unique isoforms. A novel isoform classification tool, SQANTI (https://bitbucket.org/ConesaLab/sqanti), was used to characterize isoforms with over 30 descriptors. Here we provide sequences of the resulting high quality, full-length isoforms and the annotation using the current rheMac8 assembly. SQANTI classification data is also provided for both rhesus macaque and human.
A publication detailing this study is under review. Iso-Seq raw reads have been deposited into the NCBI Sequence Read Archive (SRA), and it will become immediately public once the manuscript is accepted. The processed isoform data set and associated annotation is available here.
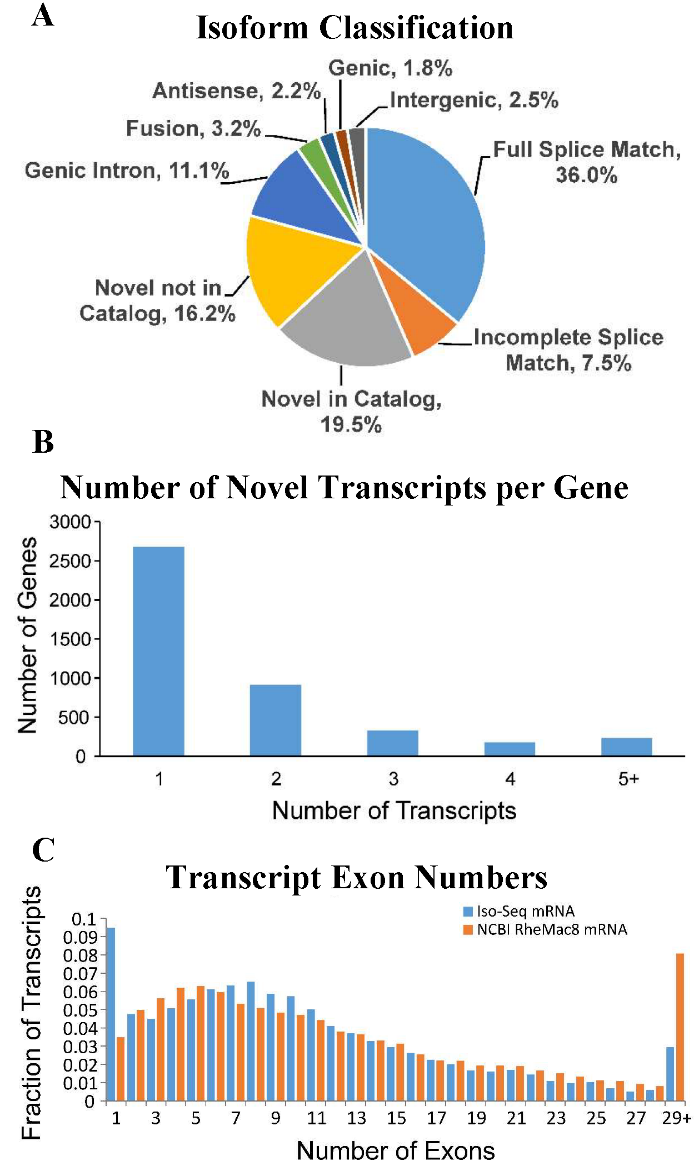 |
A) SQANTI classification of transcript isoforms indicates large fraction of novel isoforms (NIC, NNIC). B) Significant fraction of identified annotated genes were shown to have at least one novel isoform. C) Distribution of transcript exon numbers greatly resembles that of the NCBI RheMac8 annotation. |
